Data source
These charts are drawn using data published by the Official Statistics Portal (OSP) on their COVID-19 open data site, along with the annual population counts for Lithuanian municipalities, also published by the OSP.
The R markdown source is available as a github repo.
Show code
age_bands_municipalities <- tibble(lt_aggregate) %>%
# select(-object_id) %>%
mutate(date = as_date(date)) %>%
group_by(municipality_name, date, age_gr) %>%
summarise(across(where(is.numeric), ~ sum(.x, na.rm=TRUE))) %>%
ungroup()
age_bands <- age_bands_municipalities %>%
group_by(date, age_gr) %>%
summarise(across(where(is.numeric), ~ sum(.x, na.rm=TRUE)))
natl_age_data <- lt_age_sex_data %>%
filter( location == "Total" ) %>%
select(-location) %>%
pivot_longer(
cols = !"total",
values_to = "count",
names_to = "age_range") %>%
mutate_if(is.character, str_remove_all, pattern = "\\d+[^\\d]") %>%
mutate(age_range = as.numeric(age_range)) %>%
mutate(age_range = replace_na(age_range, 85)) %>%
mutate(cohort = cut(age_range,
c(0, 9, 19, 29, 39, 49, 59, 69, 79, 89 ),
c("0-9", "10-19", "20-29", "30-39", "40-49", "50-59", "60-69", "70-79", "80+"),
include.lowest = TRUE)) %>%
select(-age_range,-total) %>%
group_by(cohort) %>%
summarise(count = sum(count))
lt_age_data <- lt_age_sex_data %>%
filter(grepl("mun.$", location)) %>%
pivot_longer(
cols = !c("location","total"),
values_to = "count",
names_to = "age range") %>%
mutate_if(is.character, str_replace_all, pattern = " mun.", replacement = "")
age_band_factors <- age_bands %>%
mutate(cohort = case_when(
age_gr == "80-89" ~ "80+",
age_gr == "90-99" ~ "80+",
age_gr == "Centenarianai" ~ "80+",
age_gr == "Nenustatyta" ~ NA_character_,
TRUE ~ age_gr
)) %>%
filter(!is.na(cohort)) %>%
select(-age_gr) %>%
group_by(date, cohort) %>%
summarise(across(where(is.numeric), ~ sum(.x, na.rm=TRUE))) %>%
ungroup()
per_capita_rates <- left_join(age_band_factors, natl_age_data, by = c("cohort")) %>%
mutate(population = count,
cases_per_100k = new_cases / count * 100000,
deaths_all_per_mill = deaths_all / count * 1000000) %>%
select(-count)
Show code
colourCount = length(unique(age_bands$age_gr))
getPalette = colorRampPalette(brewer.pal(8, "Set2"))
age_bands %>%
group_by(age_gr) %>%
mutate(cases_7d = zoo::rollmean(new_cases,k=7, fill=NA) ) %>%
ungroup() %>%
filter(date > ymd("2021-08-01")) %>%
ggplot(aes(x = date, y=cases_7d, fill=age_gr)) +
theme_minimal() +
#geom_col(width=1, position = position_stack(reverse = TRUE)) +
geom_area() +
#scale_fill_brewer(palette = "Set2") +
scale_fill_viridis_d(
name = "Age cohort",
#option = "inferno",
breaks = c("0-9", "10-19", "20-29", "30-39", "40-49", "50-59", "60-69",
"70-79", "80-89", "90-99", "Centenarianai", "Nenustatyta"),
labels = c("0-9", "10-19", "20-29", "30-39", "40-49", "50-59", "60-69",
"70-79", "80-89", "90-99", "100+", "Unknown"),
direction = 1
) +
scale_y_continuous(sec.axis = dup_axis()) +
labs(title="Lithuania - COVID-19 cases by age group",
subtitle="7 day rolling average",
y="New cases",
x="Date",
caption=caption_text) +
scale_x_date()
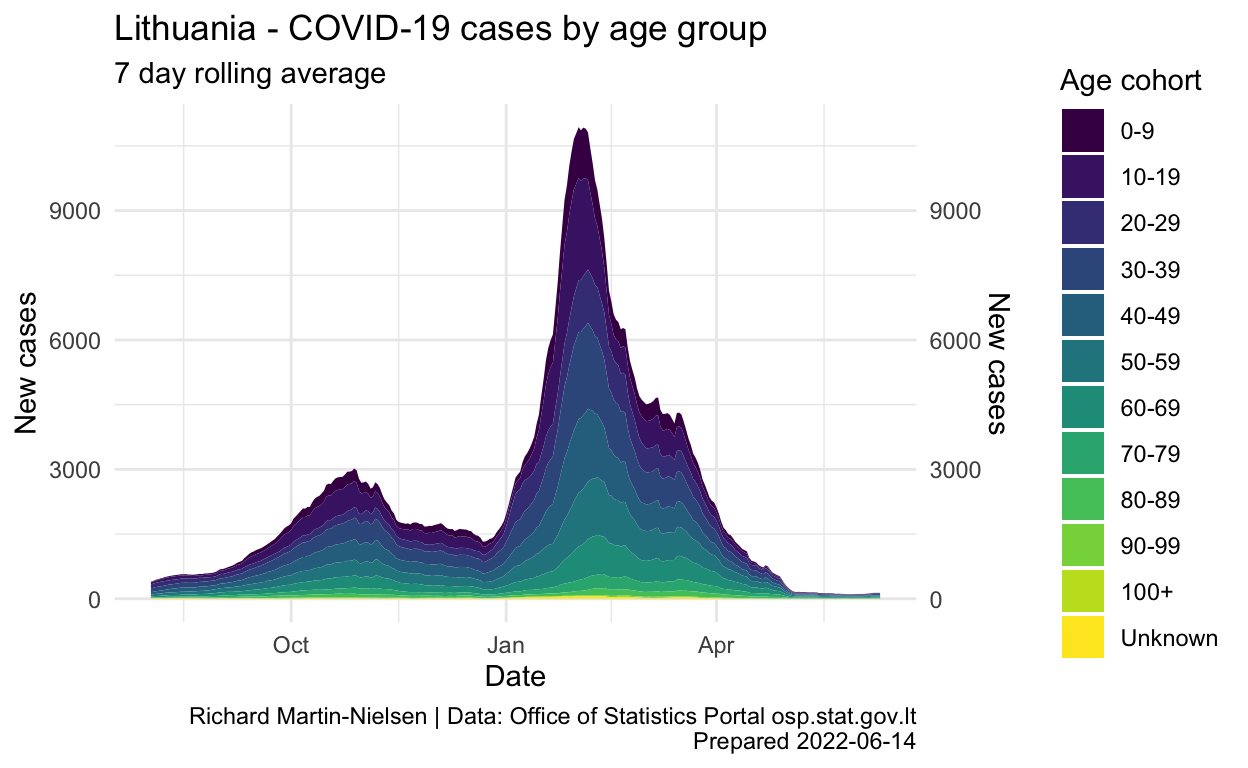
These charts are inspired by the narrow age cohort graph given by the OSP on their pandemic illustrations page:

Because the age cohorts given in the two sources used do not align, when calculating COVID rates relative to age cohorts, a smaller number of larger cohorts is used. It is also possible to extend the graph further back into 2021.
Show code
pc_rate_14d <-
per_capita_rates %>%
group_by(cohort) %>%
mutate(pc_cases_14d = zoo::rollsum(cases_per_100k,k=14, fill=NA, align="right") ) %>%
ungroup() %>%
filter(date >= ymd("2021-09-01"))
ntl_rate_14d <- per_capita_rates %>%
group_by(date) %>%
summarise(across(where(is.numeric), ~ sum(.x, na.rm=TRUE))) %>%
mutate(cases_per_100k = new_cases / population * 100000,
deaths_all_per_mill = deaths_all / population * 1000000) %>%
mutate(pc_cases_14d = zoo::rollsum(cases_per_100k,k=14, fill=NA, align="right") ) %>%
filter(date >= ymd("2021-09-01"))
pc_rate_14d %>%
ggplot(aes(x = date, y=pc_cases_14d, colour=cohort)) +
theme_minimal() +
theme( legend.position = "none") +
geom_line(size=1) +
geom_line(data = ntl_rate_14d, aes(x=date, y=pc_cases_14d),
linetype = 2, colour="black") +
geom_text_repel(aes(x=date,y=pc_cases_14d,label=cohort,colour=cohort),
nudge_x=10,
direction="y",hjust="left",
data=tail(pc_rate_14d, 9)) +
geom_text_repel(aes(x=date,y=pc_cases_14d,label="National"),
colour="black",
nudge_y=1000,
hjust="right",
data=ntl_rate_14d %>% filter(date == "2022-01-01")) +
#scale_fill_brewer(palette = "Set2") +
scale_colour_viridis_d(name = "Age cohort") +
scale_y_continuous() +
labs(title="Lithuania - COVID-19 cases by age group",
subtitle="14 day cumulative per 100,000",
y="New cases",
x="Date",
caption=caption_text) +
scale_x_date(expand = expansion(add=c(0,25)))
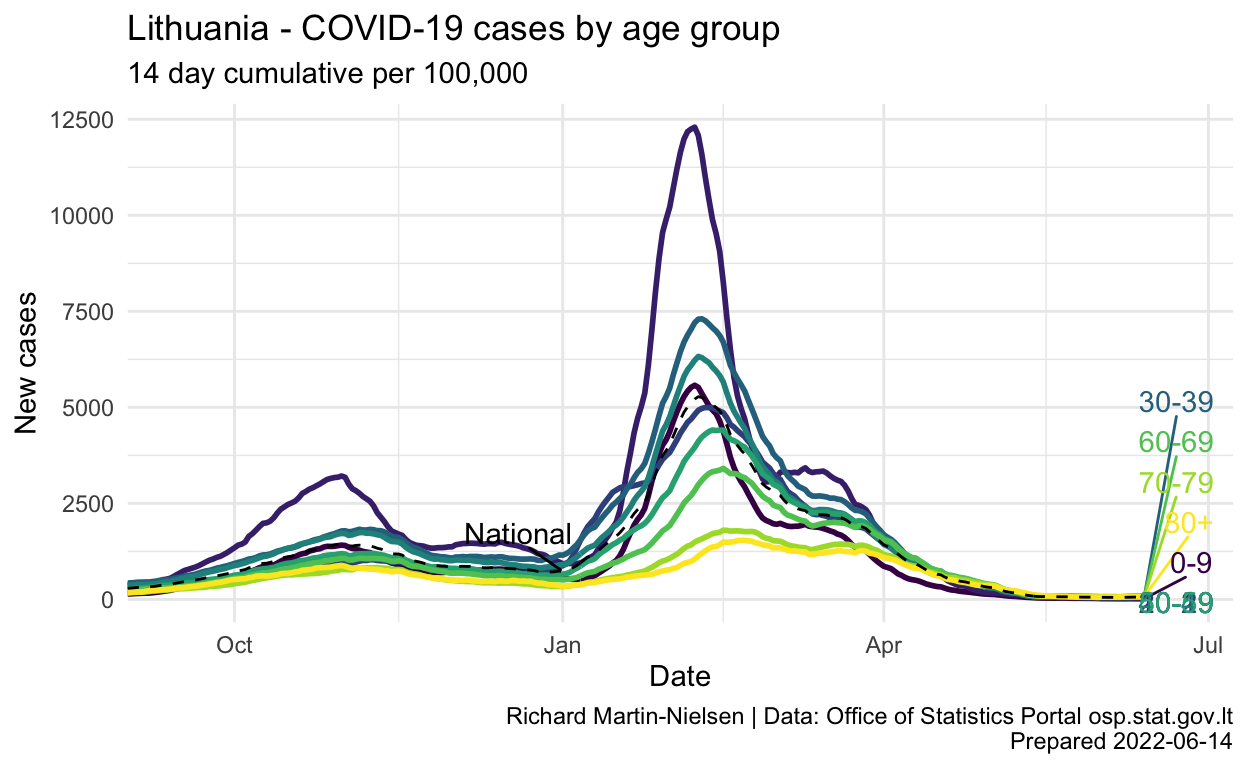
Show code
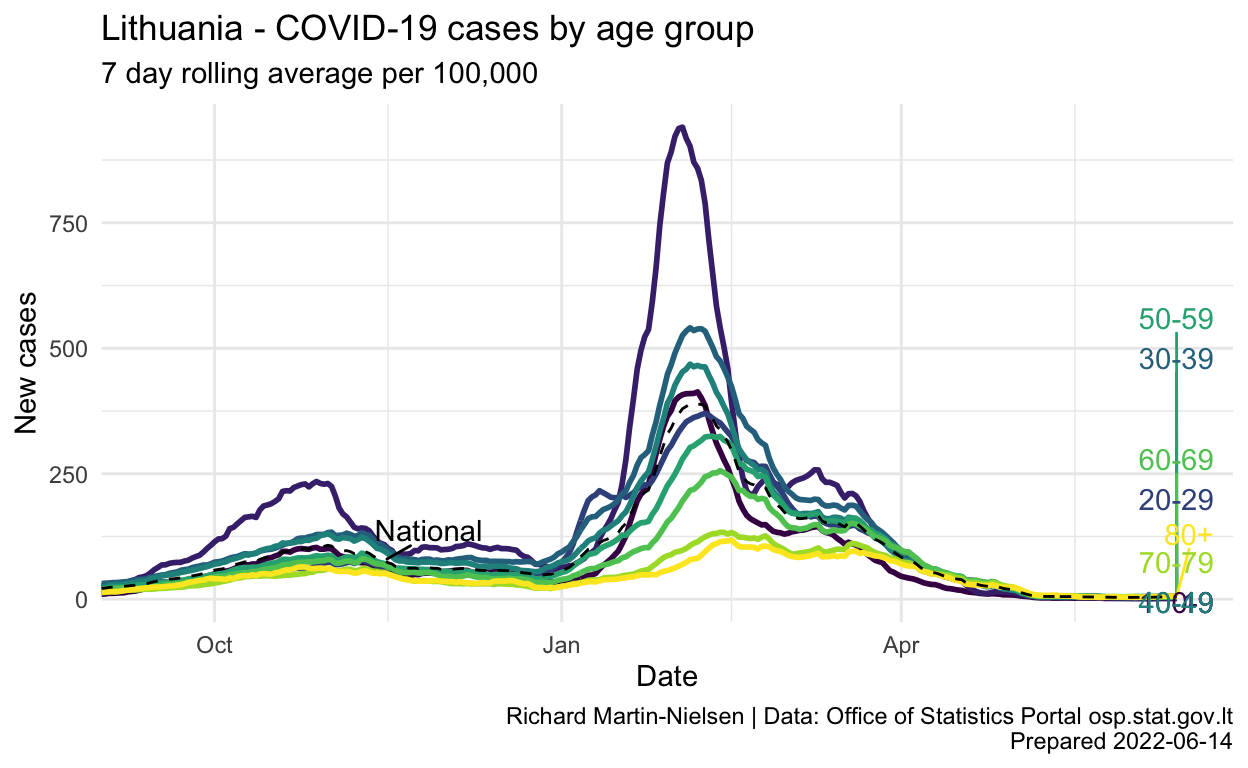
pc_rate_7d <- per_capita_rates %>%
group_by(cohort) %>%
mutate(pc_cases_7d_mean = zoo::rollmean(cases_per_100k,k=7, fill=NA, align="right") ) %>%
ungroup() %>%
filter(date >= ymd("2021-09-01"))
ntl_rate_7d <- per_capita_rates %>%
group_by(date) %>%
summarise(across(where(is.numeric), ~ sum(.x, na.rm=TRUE))) %>%
mutate(cases_per_100k = new_cases / population * 100000,
deaths_all_per_mill = deaths_all / population * 1000000) %>%
mutate(pc_cases_7d_mean = zoo::rollmean(cases_per_100k,k=7, fill=NA, align="right") ) %>%
filter(date >= ymd("2021-09-01"))
pc_rate_7d %>%
ggplot(aes(x = date, y=pc_cases_7d_mean, colour=cohort)) +
theme_minimal() +
theme( legend.position = "none") +
geom_line(size=1) +
geom_line(data = ntl_rate_7d, aes(x=date, y=pc_cases_7d_mean),
linetype = 2,
colour="black") +
geom_text_repel(aes(x=date,y=pc_cases_7d_mean,label=cohort,colour=cohort),
nudge_x=20,
direction="y",hjust="left",
data=tail(pc_rate_7d, 9)) +
geom_text_repel(aes(x=date,y=pc_cases_7d_mean,label="National"),
colour="black",
nudge_y=60,
hjust="left",
data=ntl_rate_7d %>% filter(date == "2021-11-15")) +
#scale_fill_brewer(palette = "Set2") +
scale_colour_viridis_d(name = "Age cohort") +
scale_y_continuous() +
labs(title="Lithuania - COVID-19 cases by age group",
subtitle="7 day rolling average per 100,000",
y="New cases",
x="Date",
caption=caption_text) +
scale_x_date(expand = expansion(add=c(0,15)))
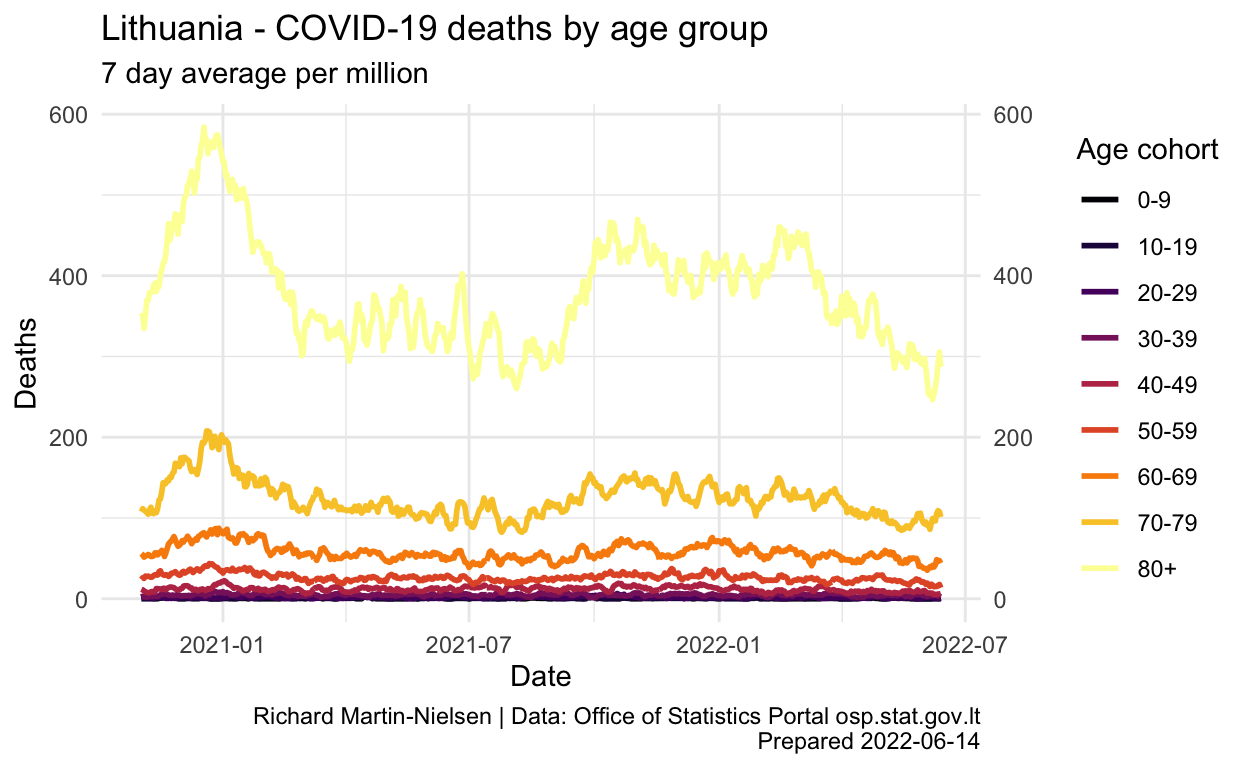
Show code
per_capita_rates %>%
group_by(cohort) %>%
mutate(pc_deaths_7d_mean = zoo::rollmean(deaths_all_per_mill,k=7, fill=0, align="right") ) %>%
ungroup() %>%
filter(date > ymd("2020-11-01")) %>%
ggplot(aes(x = date, y=pc_deaths_7d_mean, colour=cohort)) +
theme_minimal() +
geom_line(size=1) +
#scale_fill_brewer(palette = "Set2") +
scale_colour_viridis_d(name = "Age cohort", option = "inferno") +
scale_y_continuous(sec.axis = sec_axis(~ .)) +
labs(title="Lithuania - COVID-19 deaths by age group",
subtitle="7 day average per million",
y="Deaths",
x="Date",
caption=caption_text) +
scale_x_date()